Rethinking eukaryotic translation and coding potential
Historically eukaryotic mRNAs were considered to be monocistronic, containing a single functional initiation codon and it was believed that almost exclusively AUG codons are used to initiate protein synthesis. Moreover, a certain size of the translation product was considered to be necessary for biological function (initially a cut-off of 100 aminoacids was chosen). These dogmatic ideas of eukaryotic translation and mRNA coding potential have since dominated our idea of eukaryotic proteomes. A number of important findings have recently resulted in a paradigm shift. The early and reductionist ideas of eukaryotic protein synthesis are now being challenged by the growing amount of evidence that translation and coding in eukaryotes are far more plastic than previously thought.
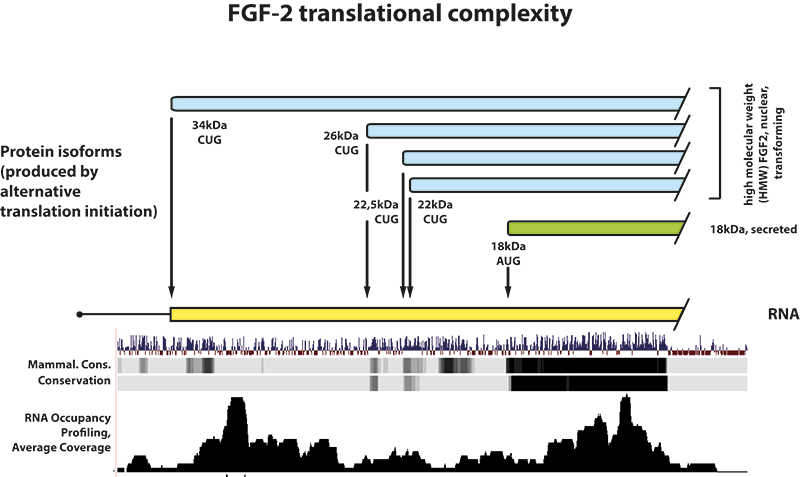
Today it is well established that even very small peptides can fulfil important biological functions such as e.g. peptide hormones and defensins in systemic homeostasis and innate immunity. Also, being first discovered as early as in the 1980ies, initiation at near-cognate start codons appears to be rather common and today is well documented for many important cellular transcripts, among them the clinically relevant c-myc, pten and fgf-2 mRNAs. Finally, there is ample evidence that a large number of eukaryotic mRNAs is indeed rather ‘polycistronic’, coding for a large (and annotated) peptide as well as additional smaller ones; and translation can occur in different and often overlapping frames.
Recently the technique of ribosomal profiling has unveiled the overwhelming complexity of eukaryotic translation initiation on a transcriptome-wide scale, providing evidence for a large number of novel coding regions and many initiation sites per RNA species. This includes the translation of many upstream open reading frames (uORFs), overlapping open reading frames, N-terminal protein extensions, and termination codon read-through, suggesting the existence of a large number of additional (poly)-peptides that are not yet annotated in the protein databases.
With financial support from the German Research Foundation (DFG) and the Federal Ministry for Education and Research (BMBF) we can now
- experimentally address translational reprogramming of stressed cells on a transcriptome-wide scale, and
- analyse initiation codon choice and its regulation in cultured models.